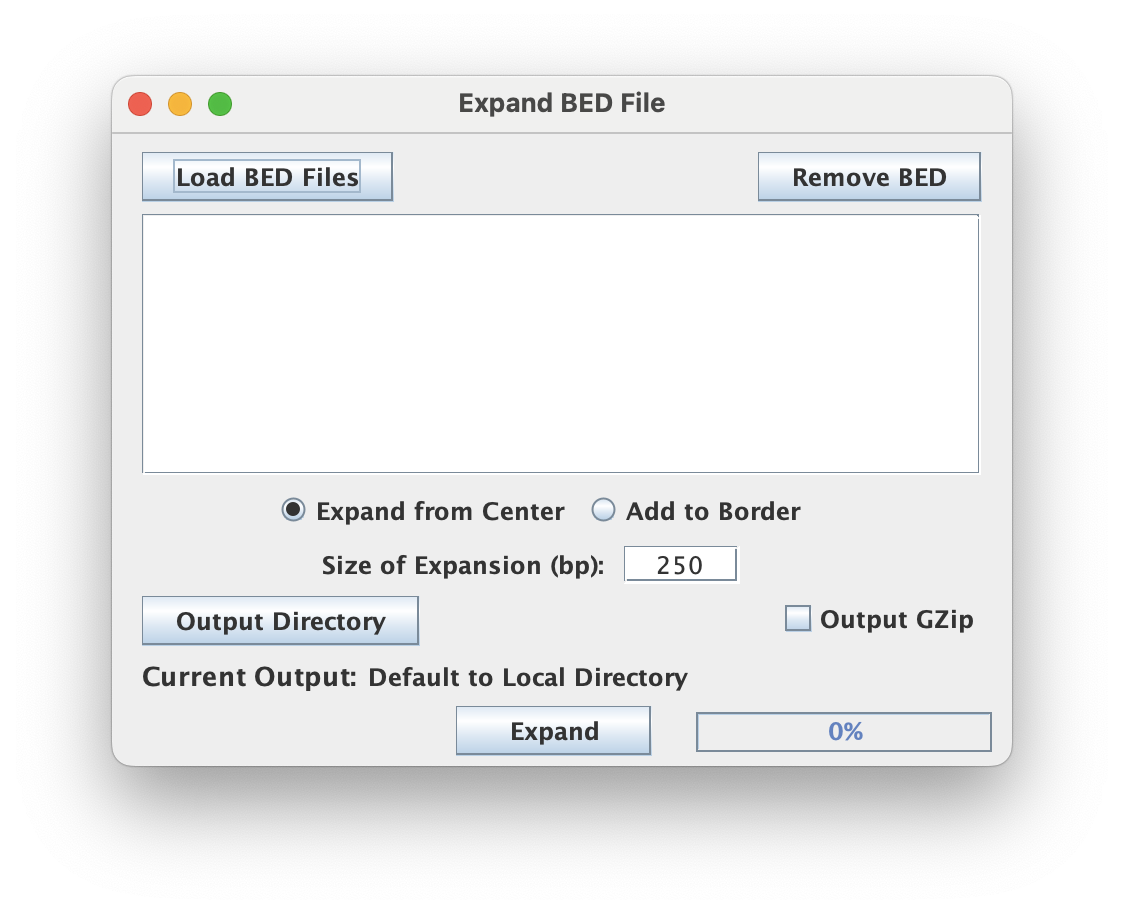
Expand BED File
Expands input BED file by adding positions to the border or around the center.
This script is related to several Bedtools functions and is typically used to expand a set of peak annotations from the center to create a set of genomic intervals of fixed size. BED files with fixed interval lengths are useful for a variety of tools including ScriptManager's Tag Pileup occupancy counts, Extract FASTA sequence for Four Color plots, and even for a number of other third-party analysis tools.
The script can also pad the intervals of BED coordinates to create intervals that aren't necessarily all of a fixed length.
Input BED files
The graphical interface restricts file selection by the .bed file extension. This tool supports batch processing of files.
Expansion strategy & size
Depending on the strategy selected, the "Size of Expansion" (in bp) can mean different things.
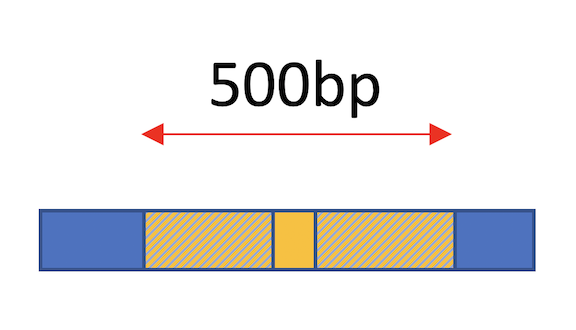
- Expand from Center: The figure at the top of the page illustrates the "expand from center" expansion strategy where the midpoint is determined by script and then the window is expanded evenly on both sides of the midpoint to the size of expansion specified by the user. This results in a BED file with intervals of a fixed length.
- Add to Border: This strategy pads both sides of the borders of the input intervals by a fixed amount specified by the user ("Size of Expansion"). Depending on whether or not the input BED file contains intervals of a fixed length, the resulting expansions will not necessarily include intervals of a fixed length.
Output format
For each input BED file, a new BED file is created with the original filename and the .bed file extension replaced with the *_<sizeofexpansion>bp.bed suffix. If the "Output GZIP" checkbox is slected, the .gz extension will also be appended.
Command Line Interface
Usage:
java -jar ScriptManager.jar coordinate-manipulation expand-bed [-hsV] [-b=<border>]
[-c=<center>] [-o=<output>] <bedFile>
Positional Input
This tool takes a single BED file for input.
Output Options
| Option | Description |
|---|---|
-o, --output | specify output filename (default name will be same as original with expansion info and .bed ext) |
-s, --stdout | output gff to STDOUT |
-z, --gzip | gzip output (default=false) |
Expansion Options
| Option | Description |
|---|---|
-c, --center | expand from center (default, at 250bp) |
-b, --border | add to border |