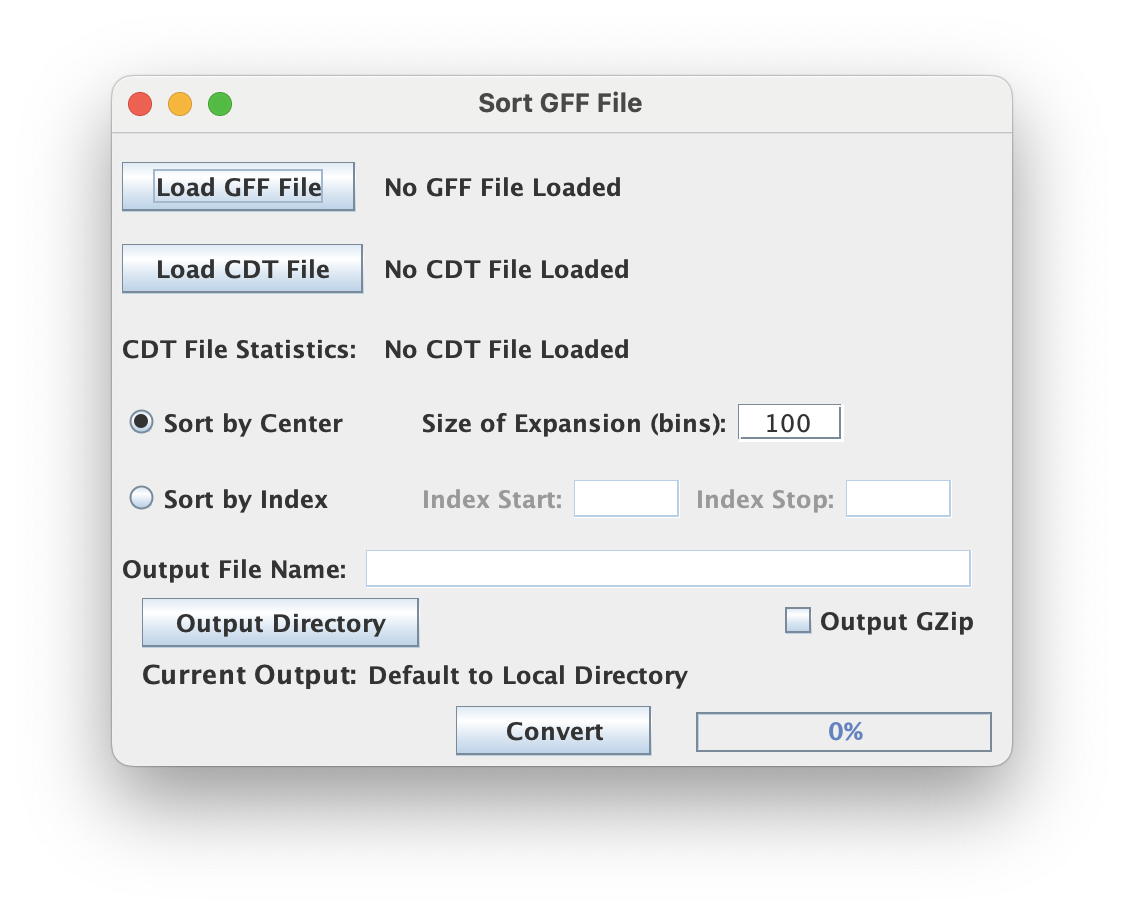
Sort GFF by CDT
Sort a CDT file and its corresponding GFF file by the total score in the CDT file across the specified interval
CDT File Statistics
CDT file statistics provide summary measures like mean, median, and standard deviation, along with distribution and clustering metrics, to help understand and analyze the genomic data's characteristics and variability.
Sorting Strategy
Depending on the strategy selected, the "Size of Expansion" (in bins) can mean different things.
- Sort by Center: This strategy sorts genomic GFF intervals according to the scores in the CDT file at the midpoint of each GFF interval.
- Sort by Index: This strategy sorts genomic GFF intervals based on scores in the GFF file at a specific index position within each BED interval.
Command Line Interface
Usage:
java -jar ScriptManager.jar coordinate-manipulation sort-gff [-hV] [-c=<center>]
[-o=<outputBasename>] [-x=<index> <index>]... <gffFile> <cdtReference>
Positional Input
| Input | Description |
|---|---|
<gffFile> | the GFF file to sort |
<cdtReference> | the reference CDT file to sort the input by |
Output Options
| Option | Description |
|---|---|
-o, --output=<outputBasename> | specify output file basename (no .cdt/.gff extension, script will add that) |
-z, --gzip | gzip output (default=false) |
Sort Options
These options indicate which windows to sort the files by (choose one).
| Option | Description |
|---|---|
-c, --center=<center> | sort by center on the input size of expansion in bins (default=100) |
-x, --index=<index> <index> | sort by index from the specified start to the specified stop (0-indexed and half-open interval) |