Random Coordinate Generator
Generate random BED coordinates based on reference genome.
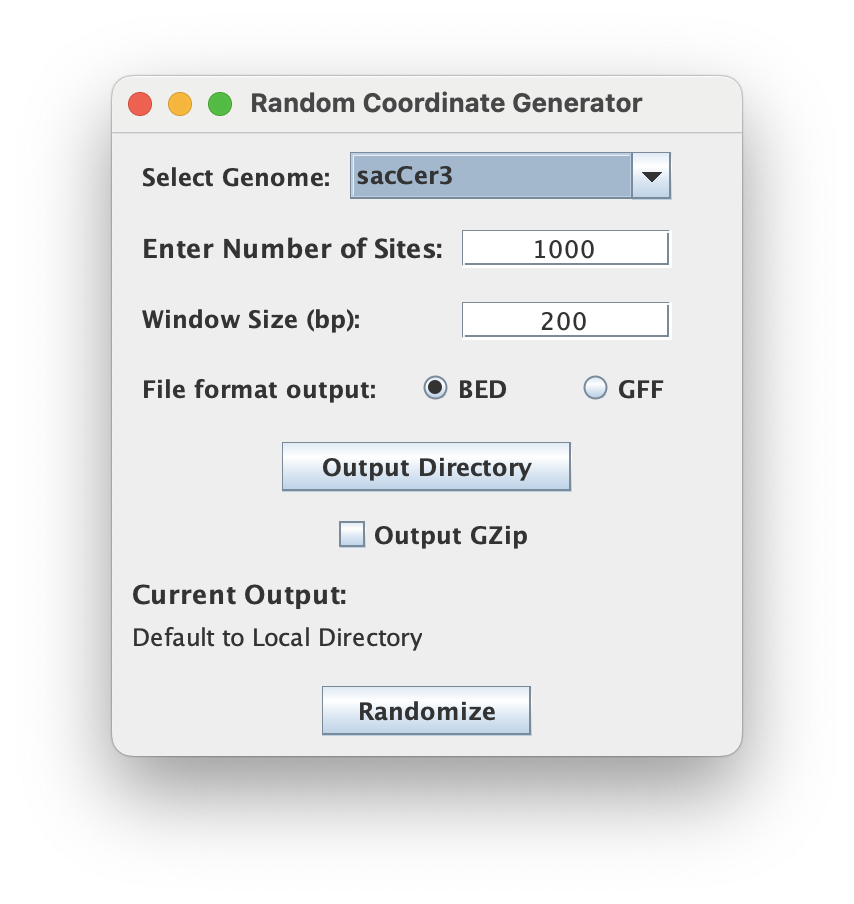
Select Genome
Select a reference genome from the provided dropdown menu. The user can customize the number of sites and the window size generated by the tool.
Calculation Options
The 'Number of Sites' specifies the total number of random genomic regions to be generated. The 'Window Size' defines the length of each random genomic region in base paris (bp).
Command Line Interface
Usage:
java -jar ScriptManager.jar peak-analysis rand-coord [-fhV] [-n=<numSites>]
[-o=<output>] [-w=<window>] <genome>
| Input | Description |
|---|---|
<genome> | reference genome sacCer3 | sacCer3_cegr | hg38 | hg38_contigs | hg19 | hg19_contigs | mm10 |
Output Options
| Option | Description |
|---|---|
-o, --output=<output> | specify output directory (name will be random_coordinates_<genome>_<window>bp.<ext>) |
-z, --gzip | gzip output (default=false) |
-f, --gff | file format output as GFF (default format as BED) |
-n, --num-sites=<numSites> | number of sites (default=1000) |
-w, --window=<window> | window size in bp (default=200) |